Using the five methods described in Technical Note: The Four Winds of Microbiome Analysis, I ran these method on all of the data on the citizen science site of Microbiome Prescription testing for all symptoms that have been self-reported from users of Ombre Labs and Biomesight retail microbiome tests. The data from each lab was done is insolation (you cannot mix data from different processions flows, see The taxonomy nightmare before Christmas… for how the results from the same FASTQ files are reported by 4 different processing flows).
My criteria for deeming a genus significant was:
- At least one method reported P < 0.01
- At least two methods reported P < 0.05
The 2 @ P < 0.05 is a bit of shooting from the hip; I expect some correlation between methods but not sufficient to have that adjusted P value to be outside of the range 0.0025 and 0.01. Looking at the statistics on significant genus, we found the 2 @ P < 0.05 produce only a small contributions,
See Citizen Science Symptoms To Genus Special Studies
When I looked at associations for autism, I noticed a striking contrast between the two most common labs.
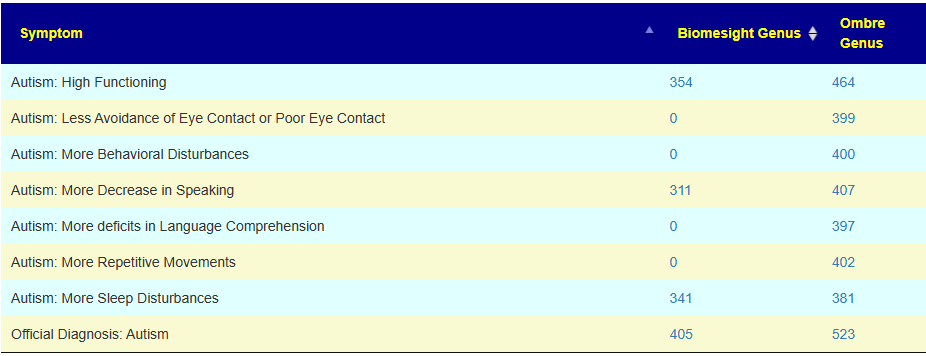
There are two possible causes:
- Less annotated samples on Biomesight uploads
- The algorithm being used on Biomesight is a poor match for the RNA used in 16s that are associated with autism (and other neurocognitive issues).
Looking at some other neurocognitive issues, we see the same pattern — Ombre identifies more significant genus.

How to Fix This Issue
The first issue may disappear if all biomesight samples with autism are annotated. HINT HINT
The second issue would be addressed by having the 16s FastQ files processed by OmbreLabs (At one time they offered that for free).
Example of Using
In the sample below, we see for Bifidobacterium that average amount is 2.6 x of the values of people without this symptom. The percentile rankings are 36% higher, and this genus is seen 3% more often.
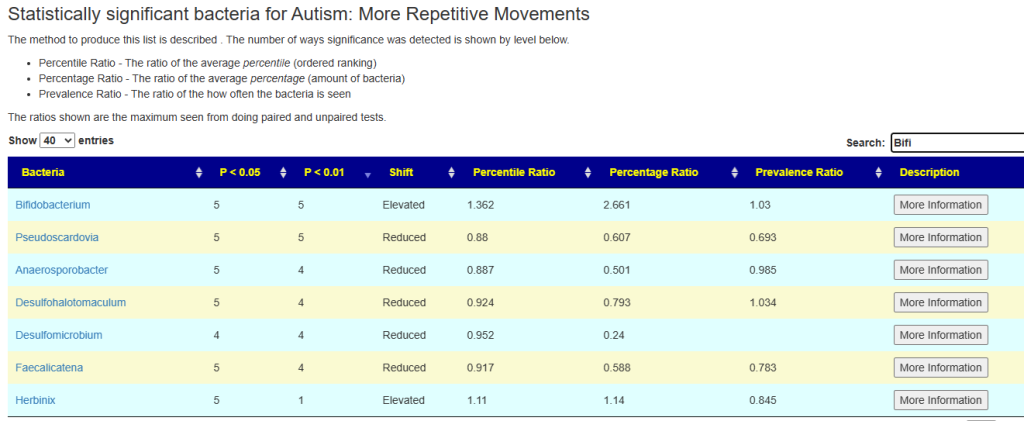
Pending Work
Integrating this data with the algorithms on Microbiome Prescription to generate suggestions to reduce these shifts.
Also see Technical Note: Yield of Applying Different Statistical Methods for more information