The interleukin 17 (IL-17) family, a subset of cytokines plays crucial roles in both acute and chronic inflammatory responses. Its intended role is against pathogens — but if it is stuck on then it becomes harmful. This can sometime happen with mis-identification of chemical signals. For details on the members of this subset, see Kyoto Encyclopedia of Genes and Genomes.
It appears to be a significant player for Autism, Interleukin-17 in Chronic Inflammatory Neurological Diseases [2020]
A reader has asked about items that are known to reduce it. The following comes from a search of PubMed
- There are a variety of prescription IL-17 inhibitors (ixekizumab, secukinumab, bimekizumab, netakimab, brodalumab) covered in this review [2021]
- An engineered Lactobacillus salivarius is described here [2017]
- Lactobacillus plantarum (LP) IS-10506 “The IL-4 and IL-17 levels were significantly lower in the probiotic than the placebo group.” [2020]
- Luteolin decreases levels [2021] (available as a supplement)
- ” The combination of L. acidophilus, vitamin B, and curcumin effectively downregulated Th17 cells and the related cytokine IL-17, thereby maintained the Treg population, ” [2020]
- Lactobacillus plantarum “pre-treatment with food-borne Lpb. plantarum significantly reduce pro-inflammatory cytokines IL-17F and IL-23 levels in inflamed NCM460 cells.” [2020]
- Lactobacillus casei Shirota (Yakult, the beverage) ” LcS significantly reduced plasma monocyte chemotactic protein-1 and, on subgroup analysis, plasma interleukin-1β (alcoholic cirrhosis), interleukin-17a and macrophage inflammatory protein-1β (non-alcoholic cirrhosis), compared with placebo.” [2020]
- The Role of Flavonoids in Inhibiting Th17 Responses in Inflammatory Arthritis [2018] provide a lot of details (including some unusual herbs and spices). Items more commonly available include:
- Apples – Procyanidins B1, B2, and C1
- Grape Seed Extract — Proanthocyanidins
- Licorice — (Glycyrrhiza glabra) I can strongly attest that it does wonders for inflammation (We use spezzatina )
- Blueberry, Raspberry, black rice, and black soybean – Anthocyanins
- Berberine “attenuating the Th17 response triggered by the B cell-activating factor” [2018]
- Astragalus “Downregulating Interleukin-17 Expression via Wnt Pathway'[2020]
- Curcumin (Turmeric) – “Curcumin mediates attenuation of pro-inflammatory interferon γ and interleukin 17 cytokine responses in psoriatic disease” [2020]
More items (with references) is listed here, this was a quick summary – that is a deeper review.
To avoid:
- Lactobacillus rhamnosus GG (LGG) ATCC 53103 “upregulated the expression of IL-17” [2020]
- “We confirm that food intake increases IL-17 expression in the mouse ileum and human blood. e. Thus, IL-17 is a gut-produced factor that is controlled by diet and modulates food intake by acting in the hypothalamus. Our findings provide the first evidence of a cytokine that is acutely regulated by food intake and plays a role in the regulation of eating.” [2020]
Bottom Line
The last citation points to the microbiome as a very significant factor for the levels of IL-17. At present, we do not know which bacteria play a role (many bacteria cannot be cultured, which limits our knowledge of what they do).
Modelling Candidate Bacteria
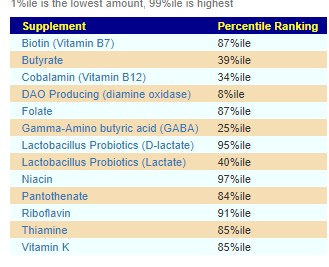
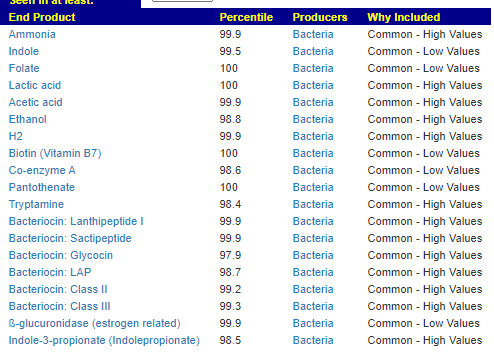
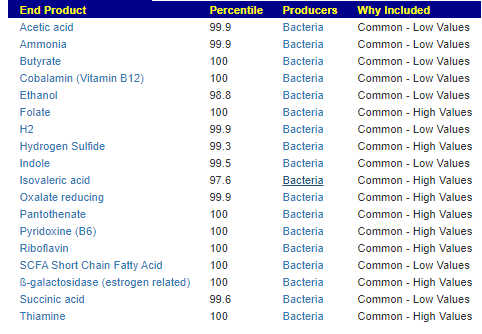
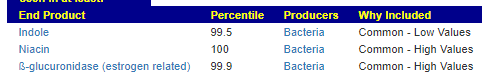
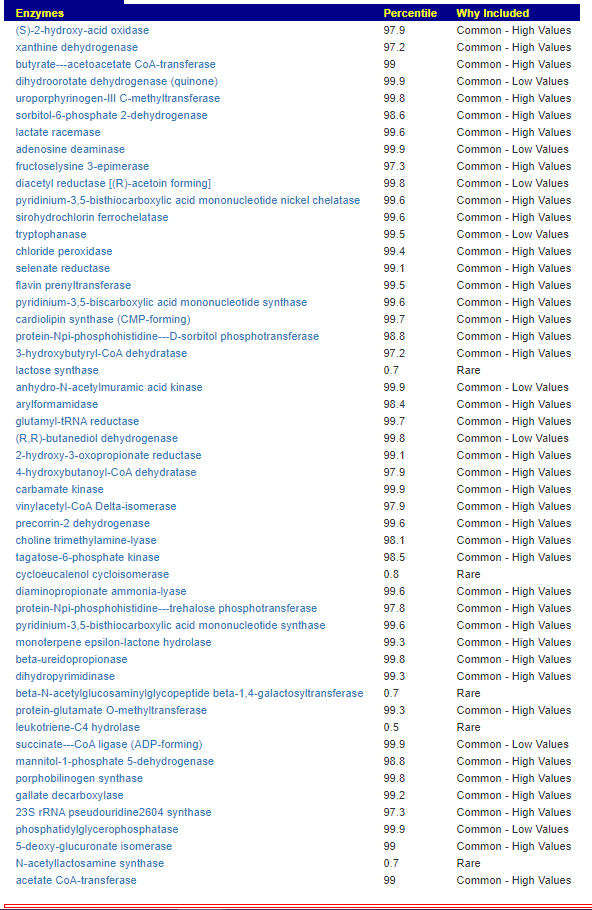
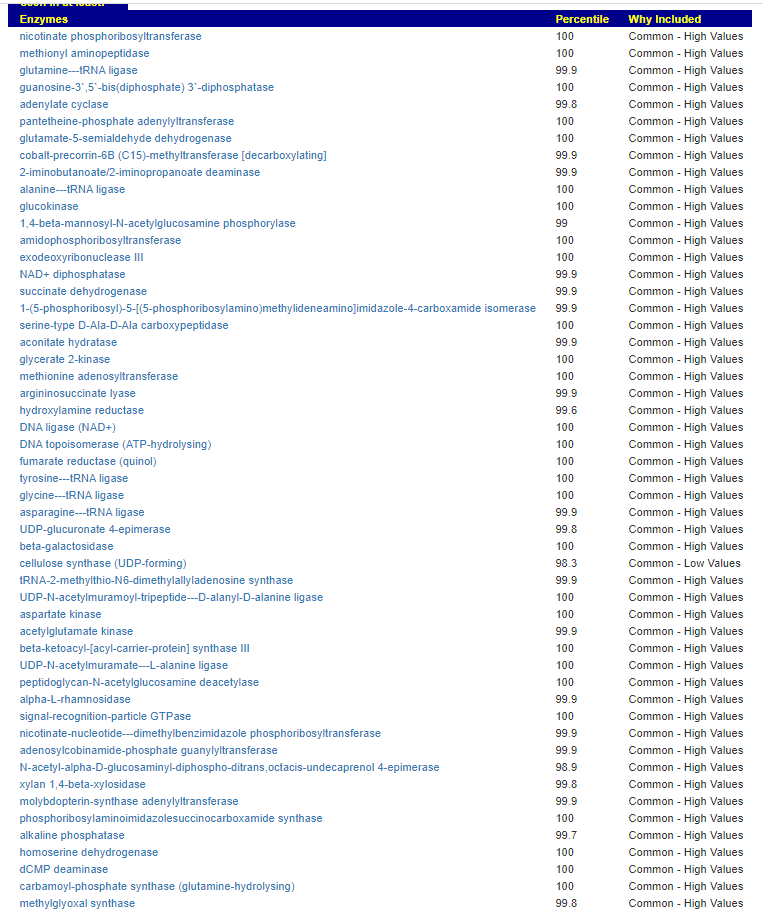
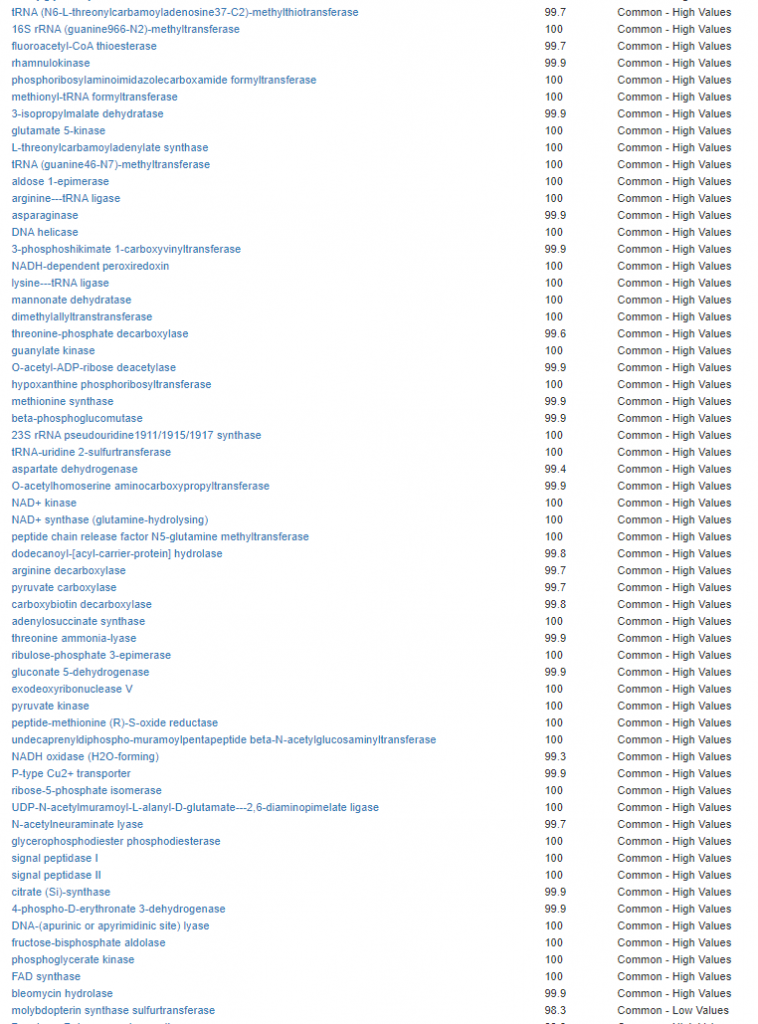
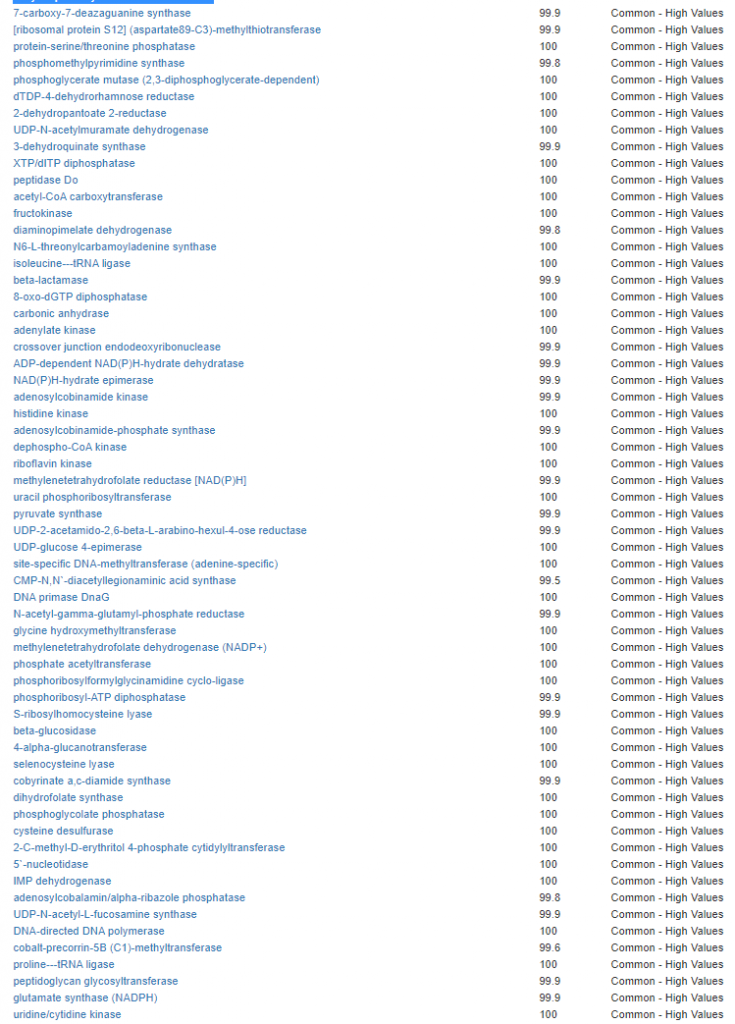
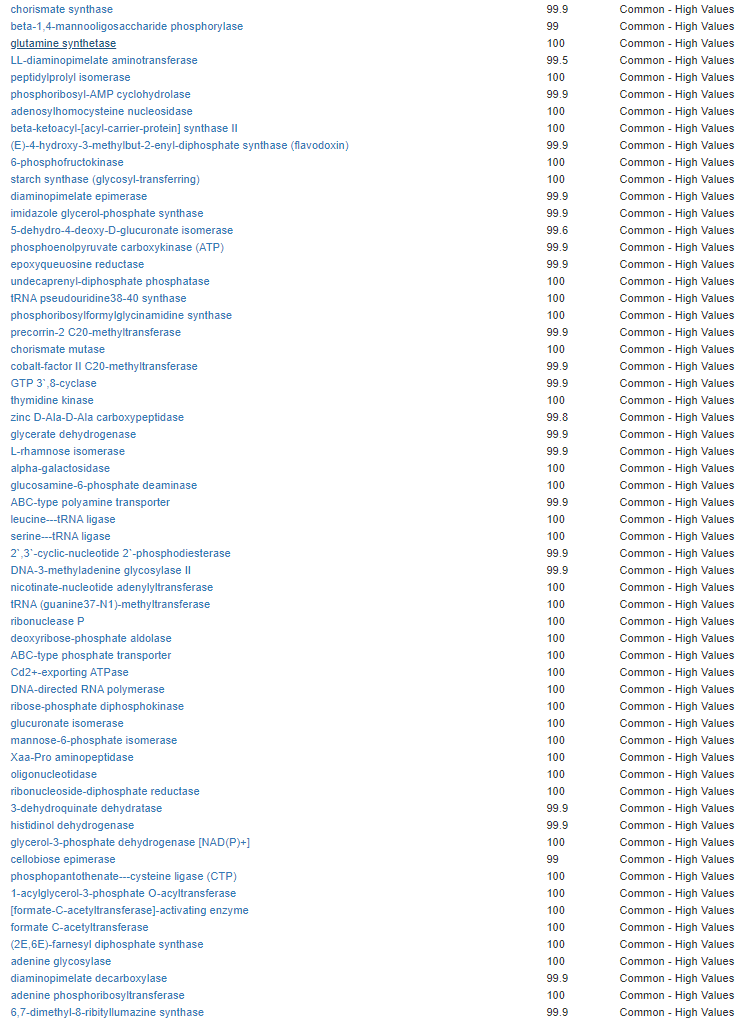
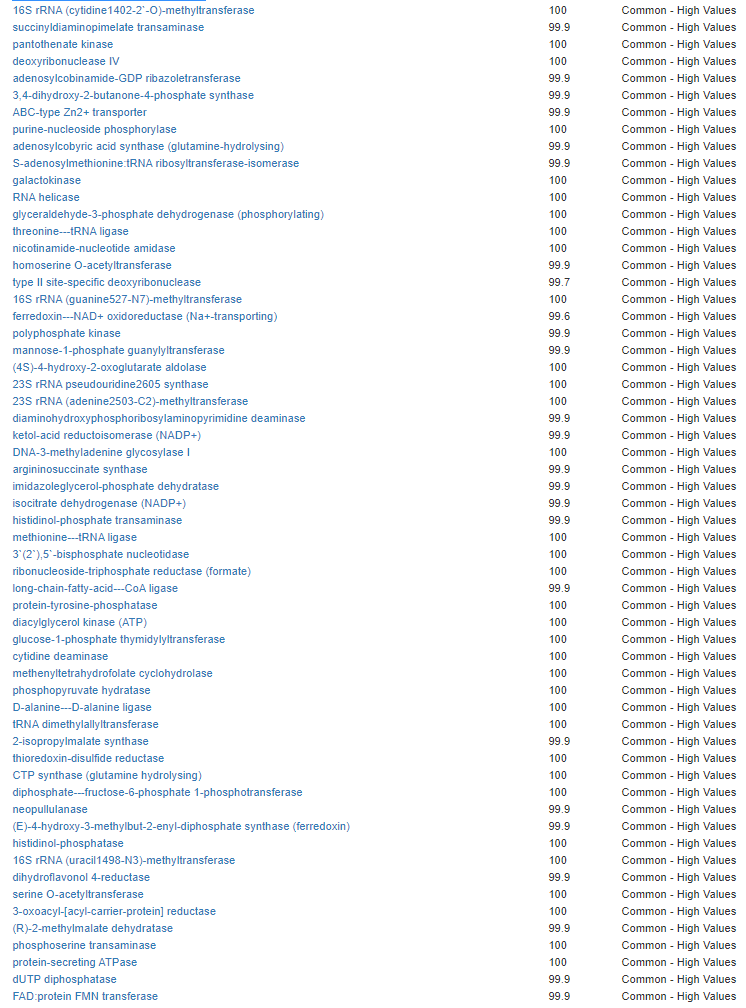
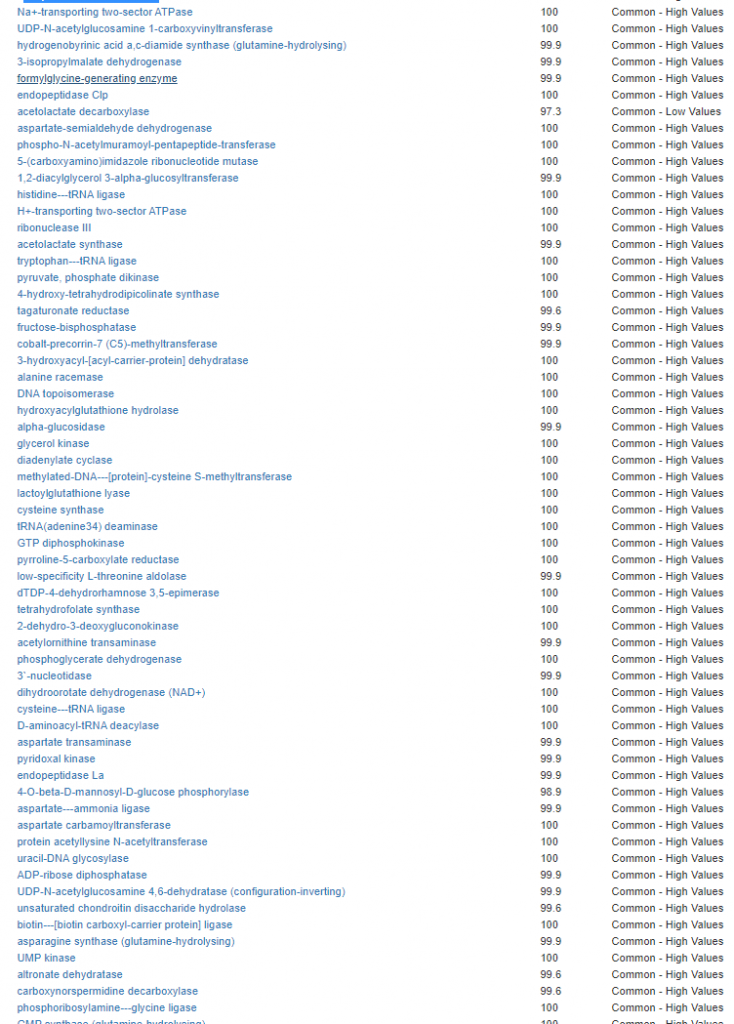
We have a list of items that reduces IL-17. We also find many of these items in our database. Thus if we look at the bacteria that are reduced by these food, we may be able to generate a candidate list of bacteria of concern.
The result is this list of significant bacteria
- Clostridiaceae (family)
- Enterobacteriaceae (family)
- Staphylococcaceae (family)
- Streptococcaceae (family)
- Clostridium (genus)
- Enterobacter (genus)
- Enterococcus (genus)
- Escherichia (genus)
- Kluyvera (genus)
- Pseudomonas (genus)
- Staphylococcus (genus)
- Streptococcus (genus)
- Enterococcus faecalis (species)
- Escherichia coli (species)
- Pseudomonas aeruginosa (species)
- Pseudomonas aeruginosa group (species)
- Staphylococcus aureus (species)
- Streptococcus mutans (species)
The strongest hint is for Staphylococcus aureus (species) which leads to this article:
- Identification of the pivotal differentially expressed genes and pathways involved in Staphylococcus aureus-induced infective endocarditis by using bioinformatics analysis [2021] … “while the pathway analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed that CDEGs were significantly enriched in the B-cell receptor, IL-17, and NF-kappa B signaling pathways.”
- Specific IgA and CLA + T-Cell IL-17 Response to Streptococcus pyogenes in Psoriasis [2020]
- “The highest IL-17a level occurred in patients with chronic plus the acute isolation of Pseudomonas aeruginosa. ” [2020]
So we have a full cycle… items shown to reduce IL-17 also are items that reduce a list of bacteria. Checking those bacteria, we find that they are associated with high IL-17 levels.
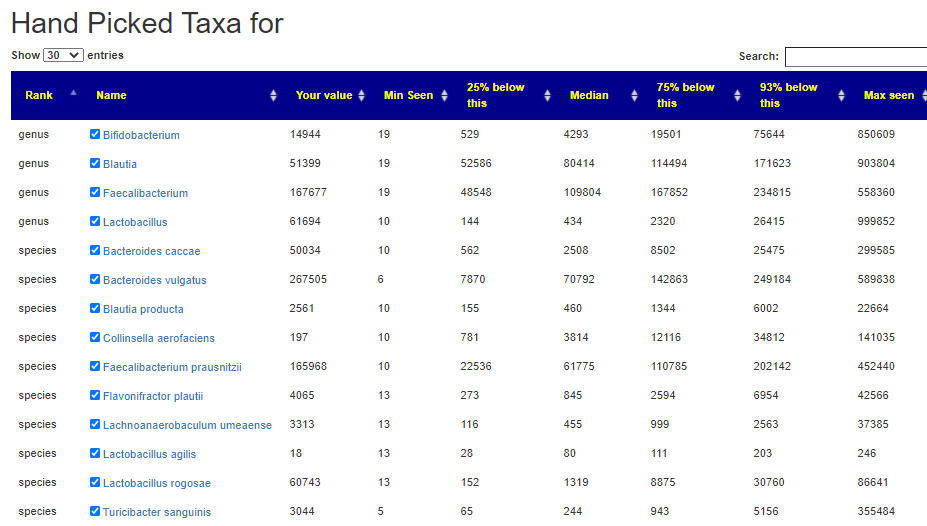
Consequence: In asking for suggestions – you may wish to go to hand-pick bacteria and select any of those listed above.
I have a longer list by genus below that are also suspect (from strongest hint to weaker).
- Escherichia (genus)
- Pseudomonas (genus)
- Staphylococcus (genus)
- Kluyvera (genus)
- Streptococcus (genus)
- Enterobacter (genus)
- Clostridium (genus)
- Enterococcus (genus)
- Citrobacter (genus)
- Dorea (genus)
- Eubacterium (genus)
- Raoultella (genus)
- Shigella (genus)
- Bacillus (genus)
- Fenollaria (genus)
- Intestinimonas (genus)
- Caloramator (genus)
- Oscillibacter (genus)
- Gracilibacter (genus)
- Coprobacter (genus)
- Slackia (genus)
- Helicobacter (genus)
- Coprococcus (genus)
- Cronobacter (genus)
- Prevotella (genus)
- Anaerobutyricum (genus)
- Parasporobacterium (genus)
- Anaerobium (genus)
- Anaerotignum (genus)
- Fusicatenibacter (genus)
- Hespellia (genus)
- Faecalicatena (genus)
- Tyzzerella (genus)








Recent Comments